Site-based integrative analysis of biodiversity in tropical rainforests

Butterfly Diaethria clymena (Cramer, 1775). Family: Nymphalidae.© Max Barclay
Project summary
- Focus: To unearth the biodiversity of tropical rainforests in a site-based approach, using both DNA and traditional sampling methods
- Funding: The Museum
- Start date: 2013
- End date: 2016
We are assessing the complex biodiversity of tropical rainforests using innovative sequencing methods to increase the ease and rate of species identification.
Our in-depth assessment of spatially defined plots in the rainforest will also include standardised sampling methods.
Site-based analysis will enable us to compare total biodiversity and turnover in the same sites and in different locations.
By integrating the analysis of genetic and species levels, we expect to discover new emerging patterns that will help us understand the processes that drive biodiversity.
In addition, in most parts of the world the remaining primary forest sites are adjacent to areas in various states of disturbance and secondary regrowth. Studying the dynamics of change and its
Site-based approach
We are following a site-based approach, carrying out in-depth assessment of diversity at specific localities in tropical rainforests of Borneo and Latin America.
We will be comparing total diversity and turnover at study sites and using this information to establish general patterns of biodiversity.
Site-based studies have the advantage of
- simplified logistics
- repeat visits to
- conduct long-term trapping
- study seasonal change

Tropical rainforest in Panama
Sampling
We are sampling at the site using standardized protocols for
- arthropod sampling
- spatially explicit sampling
- calibrating short sampling against permanent series
- generate DNA grade specimen
Arthropod samples obtained with standard trapping methods (Malaise, flight-interception, pitfall) usually produce thousands of specimens from a complex mixture of species (‘biodiversity soup’”).
Specimen sorting and species-level identification is extremely time consuming and requires specialist expertise.

Sampling in the rainforest
Collection and storage
Dry and frozen collection workflow (unifying existing Museum collections and databases, linked to KEmu collection management system).
Metagenomics
We have adapted the metagenomics procedures used to study complex mixtures of samples in microbial communities to use mitochondrial (mt) genomes.
Only mitochondrial DNA is targeted for sequence analysis, after further enrichment with various procedures that exploit the greater AT content of mtDNA compared to nuclear DNA in insects.
Next-generation sequencing
Technology produces huge numbers of sequence reads, which permits the cost effective analysis of mtDNA even if they constitute only a small fraction of the total.
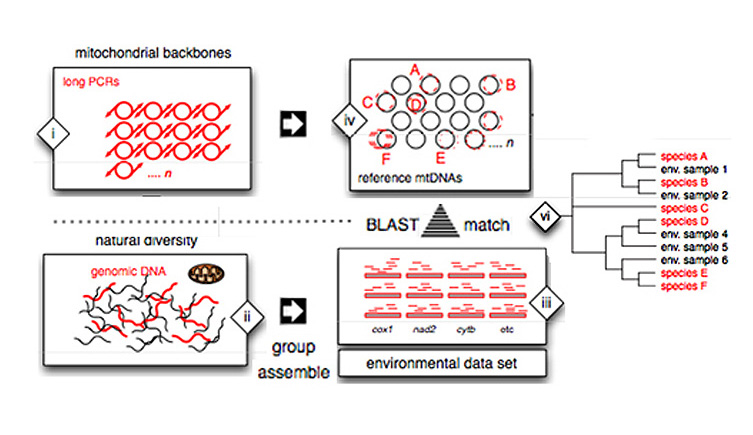
A reference database of mitochondrial genomes is generated, by long-range PCR or deep sequencing from genomic DNA. (ii) Total genomic DNA is isolated from bulk samples. (iii) Contigs are produced short sequence reads. (iv) Contigs and individual reads are aligned to reference database. (v) each sequence read is counted against the reference database, as a measure of abundance.
Statistical modelling
Our general model of biodiversity for interpreting site-based sequence data is based on stochastic (random) dispersal.
Species-genetic diversity correlation (SGDC)
When analysing a single site, neutral processes predict that total species diversity at a site (as a function of size, connectivity and age of a habitat patch) is positively correlated to the genetic diversity of each local species (as a function of population size and age).
SGDC provides a framework for testing diversity patterns at multiple hierarchical levels.

Panel A Local communities shown as white rectangles are composed of multiple species (large coloured rectangles A-D) each with multiple haplotypes (small rectangles 1-12). Note that higher species numbers are correlated with the number of genotypes per species.Species may be not shared between both communities, resulting in β-diversity. These species (A, B, D) reduce the similarity between communities. Likewise, haplotypes of shared species may contribute to turnover at the haplotype level (i.e. 7,8,9). The metacommunity is connected among sampling sites by migration m, speciation and extinction, whose rates depend on abundance of a species and connectivity.
Panel 1B. Plots of pairwise community similarity (derived from the species and haplotypes shared, as shown in Panel A), as a function of geographic distance between these communities. The example shows the greater community similarity when studied at the species level of species than haplotype level, i.e. the β-diversity is greater for haplotypes. The predicted self-similarity at both hierarchical (temporal) levels over different spatial scales is shown by the arrows. Note that one species out of 4 is shared among both communities (S=1/4) but only 1 haplotype of 12.
Museum staff
Contact us
Would you like to get involved?
We are interested in collaborations with researchers involved in studying:
- tropical ecosystems of arthropods worldwide
- agroecosystems and environmental monitoring
- application of site-based approaches of mitogenomes to non-arthropods.
Publications
Related information
Funded by
The Museum

Biodiversity research
We are creating molecular and digital tools to explore undiscovered biodiversity

Diversity and informatics research
Researching undiscovered diversity in megadiverse systems using big data

Entomology collections
Browse the oldest entomology collection in the world of over 34 million insects and arachnids